

|
| This project is collaborated between The Forsyth Institute (TFI) and The Institute for Genomic Research (TIGR), and is funded by National Institute of Dental and Craniofacial Research (NIDCR) |
|
|
|||||||||||||||||||||||||
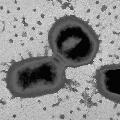
To determine the DNA sequence for the genome of P. gingivalisA random shotgun sequencing strategy will be used to determine the DNA sequence for the genome of Porphyromonas gingivalis (virulent strain W83, approximately 2.2 X 10ext6 base pairs). A 2.0 kb average insert size library will be prepared in the pUC18 vector and an 18.0 kb average insert size library in Lambda DashII. Approximately 37,000 sequences from both ends of 17,600 pUC clones and 1000 Lambda DashII clones will yield approximately 8-fold coverage of the genome. The genome sequence will be assembled from the collection of randomly sequenced fragments; sequencing gaps will be closed by primer walking on appropriate clones, and physical gaps closed by polymerase chain reaction. Editing will be done by direct inspection of the chromatograms by an experienced team and frameshift and ambiguities will be resolved using an alternative DNA sequencing chemistry. |
|||
|
This page is created and maintained by Drs. Margaret Duncan, Floyd Dewhirst, and Tsute Chen, Department of Molecular Genetics, The Forsyth Institute . Last modified on 02/20/2002 Copyright 2000, 2001, 2002 by The Forsyth Institute |